Development of a Ploidy Distinction Application: A Machine Learning Approach for Discriminating Triploid and Diploid Grass Carp (Ctenopharyngodon idella) in the Field
USGS scientists are researching a machine learning approach that can enable fisheries personnel in the field to determine the ploidy of wild-caught invasive carp.
The Science Issue and Relevance: Since being imported into the United States in 1963 to act as a biocontrol for unwanted aquatic vegetation, Grass Carp (Ctenopharyngodon idella) have been manipulated to be produced as triploids by methods such as placing fertilized eggs under pressure. Because triploid fish have three sets of chromosomes in cells, they are functionally sterile and cannot reproduce. Since the mid-1980s, the U.S. Fish and Wildlife Service (USFWS), National Triploid Grass Carp Inspection and Certification Program (USFWS; NTGCICP) has permitted and certified hatchery-produced triploids for environmental management. Despite USFWS certifications, and because of escapements or releases, diploid Grass Carp have established breeding populations throughout the Mississippi River basin, have recruited within tributaries of the Lake Erie Basin, and have been observed in Lake Michigan. Tracking of both diploid and triploid Grass Carp, as well as alerts, can be facilitated through the USGS Nonindigenous Aquatic Species database. Wild breeding populations of Grass Carp are a management concern with ecological risks to habitat and fish communities in natural systems.
States are variably permissive regarding hatchery-produced triploid Grass Carp, with all the states bordering Canada either prohibiting them or only allowing triploids. In the United States, the initiation dates for laws or policies regulating triploid Grass Carp started in 1969 in Michigan, with dates and regulations being dissimilar among the states. With no law enforcement component in NTGCICP, with states having variable prohibitions regarding diploids, and with fish crossing state boundaries, the ability of resource managers to discriminate the ploidy of invasive carp in the wild is an important tool for informing biologists’ efforts to control invasive carp spread and to reduce potential ecosystem impacts.
To assist research- and fish biologists in assessing ploidy from wild captures, and to ultimately help with management of invasive carp populations, the USFWS Fish Health Center at La Crosse, WI has been determining ploidy since 2013. This sustaining national program follows the USGS flow cytometry technology transfer of DNA content analysis using cells from Grass Carp eyeball samples. For the western United States, the USFWS Bozeman Fish Health Center, Bozeman, MT determines ploidy induction success from larval fish for hatchery-produced triploid fishes to be stocked for recreational fishing.
Although reliable and robust protocols have been developed for yielding accurate Grass Carp ploidy results in the laboratory (i.e., eyeballs, larval spawns, fish and blood), field biologists do not have a pragmatic method for discriminating triploid from diploid Grass Carp. Currently, eyeballs or blood collected in the field are shipped to distant labs for analyses. A field-friendly method could support law enforcement actions in defense of the Lacey Act, help managers obtain real-time data to apply to immediate management actions as needed, and could inform long-term management strategies.
Methodology for Addressing the Issue: A machine learning approach is being researched as a way for fisheries personnel in the field to determine the ploidy of wild-caught invasive carp. The use of machine learning in biomedical imaging accumulates substantial volumes of data via digital images, applies computational tools for processing these data, and can combine images from various microscopes by enhancing and equalizing image resolution. Thus, a computational model was trained with selected brightfield microscopic images of whole blood smears made from known diploid and triploid Grass Carp. Digital images were generated by using two types of microscopes: (1) a laboratory-dedicated, high caliber scope, and (2) a portable microscope meant for use in the field.
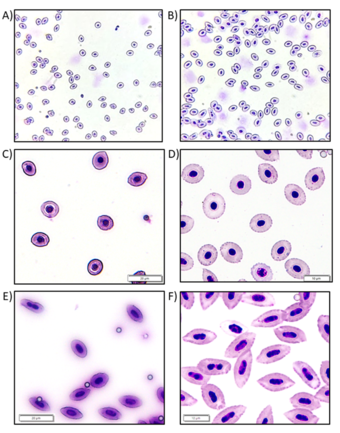
In triploid and polyploid fish, red blood cell (RBC) morphometrics are different than those from reproductive diploid fish, as documented by microscopy and flow cytometry studies. Triploids display abnormally shaped nuclei (e.g., hourglass, and not completely oval), and lengthened nuclei and total cell lengths (Fig. 1). Diploid broodstock from CERC and WARC-maintained triploid Grass Carp were bled, blood smears were hematologically stained, and microscopic imaging was performed with a field microscope attached to a cell phone adaptor at 600x total magnification, and with a standard laboratory microscope at 400x and 600x (Figs. 2 and 3). Polystyrene beads with a 3.1 µm diameter were included as an internal size control to account for possible variability in blood sample handling (Fig. 2).
After USGS Cloud Hosting Solutions personnel normalized the microscopic photographs, an open-source “You Only Look Once” (YOLO) family of image object detection computer algorithms was applied to classify the 600 images. An accuracy of 94.8% was achieved.
Future Steps: Further training photographs could be taken so to refine the model for producing a final algorithm for establishing this Proof of Concept to ultimately engineer and deploy a Ploidy Distinction Application (App) for resource manager and field biologist use. To enable migration of the model for the Ploidy Distinction App software and field application development, further technology and quick response opportunities will be investigated. The ubiquitous availability of smartphones, with integrated cameras and communication capabilities, makes them an ideal platform for field use in resource-limited settings such as along riverbanks. We envision the Ploidy Distinction App will be adaptable for multiple field microscope types and cell phone models, with the data easily communicable to partners.

A Website Tool for Predicting Triploidy in Larval Fish Spawns
Determining the Ploidy and Resultant Reproductive Capability of Artificially Spawned and Wild Caught Invasive Carp
Non-USGS related publications:
Krynak et al. 2015. https://doi.org/10.1007/s10530-015-0856-9
Okomoda et al. 2018. https://doi.org/10.1007/s00580-017-2589-x
Defensible standardized ploidy assessments for Grass Carp (Ctenopharyngodon idella, Cyprinidae) intercepted from the commercial supply chain Defensible standardized ploidy assessments for Grass Carp (Ctenopharyngodon idella, Cyprinidae) intercepted from the commercial supply chain
USGS scientists are researching a machine learning approach that can enable fisheries personnel in the field to determine the ploidy of wild-caught invasive carp.

The Science Issue and Relevance: Since being imported into the United States in 1963 to act as a biocontrol for unwanted aquatic vegetation, Grass Carp (Ctenopharyngodon idella) have been manipulated to be produced as triploids by methods such as placing fertilized eggs under pressure. Because triploid fish have three sets of chromosomes in cells, they are functionally sterile and cannot reproduce. Since the mid-1980s, the U.S. Fish and Wildlife Service (USFWS), National Triploid Grass Carp Inspection and Certification Program (USFWS; NTGCICP) has permitted and certified hatchery-produced triploids for environmental management. Despite USFWS certifications, and because of escapements or releases, diploid Grass Carp have established breeding populations throughout the Mississippi River basin, have recruited within tributaries of the Lake Erie Basin, and have been observed in Lake Michigan. Tracking of both diploid and triploid Grass Carp, as well as alerts, can be facilitated through the USGS Nonindigenous Aquatic Species database. Wild breeding populations of Grass Carp are a management concern with ecological risks to habitat and fish communities in natural systems.
States are variably permissive regarding hatchery-produced triploid Grass Carp, with all the states bordering Canada either prohibiting them or only allowing triploids. In the United States, the initiation dates for laws or policies regulating triploid Grass Carp started in 1969 in Michigan, with dates and regulations being dissimilar among the states. With no law enforcement component in NTGCICP, with states having variable prohibitions regarding diploids, and with fish crossing state boundaries, the ability of resource managers to discriminate the ploidy of invasive carp in the wild is an important tool for informing biologists’ efforts to control invasive carp spread and to reduce potential ecosystem impacts.
To assist research- and fish biologists in assessing ploidy from wild captures, and to ultimately help with management of invasive carp populations, the USFWS Fish Health Center at La Crosse, WI has been determining ploidy since 2013. This sustaining national program follows the USGS flow cytometry technology transfer of DNA content analysis using cells from Grass Carp eyeball samples. For the western United States, the USFWS Bozeman Fish Health Center, Bozeman, MT determines ploidy induction success from larval fish for hatchery-produced triploid fishes to be stocked for recreational fishing.
Although reliable and robust protocols have been developed for yielding accurate Grass Carp ploidy results in the laboratory (i.e., eyeballs, larval spawns, fish and blood), field biologists do not have a pragmatic method for discriminating triploid from diploid Grass Carp. Currently, eyeballs or blood collected in the field are shipped to distant labs for analyses. A field-friendly method could support law enforcement actions in defense of the Lacey Act, help managers obtain real-time data to apply to immediate management actions as needed, and could inform long-term management strategies.

Methodology for Addressing the Issue: A machine learning approach is being researched as a way for fisheries personnel in the field to determine the ploidy of wild-caught invasive carp. The use of machine learning in biomedical imaging accumulates substantial volumes of data via digital images, applies computational tools for processing these data, and can combine images from various microscopes by enhancing and equalizing image resolution. Thus, a computational model was trained with selected brightfield microscopic images of whole blood smears made from known diploid and triploid Grass Carp. Digital images were generated by using two types of microscopes: (1) a laboratory-dedicated, high caliber scope, and (2) a portable microscope meant for use in the field.
In triploid and polyploid fish, red blood cell (RBC) morphometrics are different than those from reproductive diploid fish, as documented by microscopy and flow cytometry studies. Triploids display abnormally shaped nuclei (e.g., hourglass, and not completely oval), and lengthened nuclei and total cell lengths (Fig. 1). Diploid broodstock from CERC and WARC-maintained triploid Grass Carp were bled, blood smears were hematologically stained, and microscopic imaging was performed with a field microscope attached to a cell phone adaptor at 600x total magnification, and with a standard laboratory microscope at 400x and 600x (Figs. 2 and 3). Polystyrene beads with a 3.1 µm diameter were included as an internal size control to account for possible variability in blood sample handling (Fig. 2).
After USGS Cloud Hosting Solutions personnel normalized the microscopic photographs, an open-source “You Only Look Once” (YOLO) family of image object detection computer algorithms was applied to classify the 600 images. An accuracy of 94.8% was achieved.
Future Steps: Further training photographs could be taken so to refine the model for producing a final algorithm for establishing this Proof of Concept to ultimately engineer and deploy a Ploidy Distinction Application (App) for resource manager and field biologist use. To enable migration of the model for the Ploidy Distinction App software and field application development, further technology and quick response opportunities will be investigated. The ubiquitous availability of smartphones, with integrated cameras and communication capabilities, makes them an ideal platform for field use in resource-limited settings such as along riverbanks. We envision the Ploidy Distinction App will be adaptable for multiple field microscope types and cell phone models, with the data easily communicable to partners.

A Website Tool for Predicting Triploidy in Larval Fish Spawns
Determining the Ploidy and Resultant Reproductive Capability of Artificially Spawned and Wild Caught Invasive Carp
Non-USGS related publications:
Krynak et al. 2015. https://doi.org/10.1007/s10530-015-0856-9
Okomoda et al. 2018. https://doi.org/10.1007/s00580-017-2589-x



