This a version of the logo for the Python Hyperspectral Analysis Tool (PyHAT). It is intended for use in info boxes on the USGS website. The spectrum in the graphic is a laser induced breakdown spectroscopy spectrum, plotted on a logarithmic y axis to emphasize weaker emission peaks.
Ryan Bradley Anderson (Former Employee)
Science and Products
Terrestrial Remote Sensing Data Ingestion with PyHAT (Python Hyperspectral Analysis Tool)
Python Hyperspectral Analysis Tool (PyHAT)
Digital Terrain Models for Mars Sinuous Ridges and Alluvial Fans Projects Digital Terrain Models for Mars Sinuous Ridges and Alluvial Fans Projects
This a version of the logo for the Python Hyperspectral Analysis Tool (PyHAT). It is intended for use in info boxes on the USGS website. The spectrum in the graphic is a laser induced breakdown spectroscopy spectrum, plotted on a logarithmic y axis to emphasize weaker emission peaks.
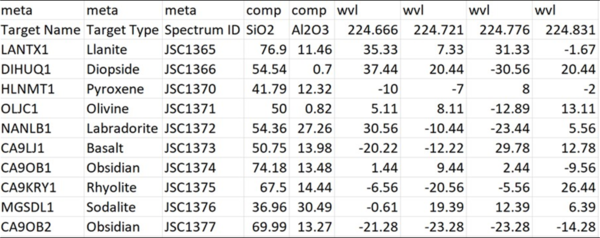
Python Hyperspectral Analysis Tool (PyHAT) Data Format Example
Python Hyperspectral Analysis Tool (PyHAT) Data Format ExampleScreenshot showing the simple data format used by the Python Hyperspectral Analysis Tool (PyHAT). Spectra are stored in rows of the table, along with their associated metadata and compositional information.
Python Hyperspectral Analysis Tool (PyHAT) Data Format Example
Python Hyperspectral Analysis Tool (PyHAT) Data Format ExampleScreenshot showing the simple data format used by the Python Hyperspectral Analysis Tool (PyHAT). Spectra are stored in rows of the table, along with their associated metadata and compositional information.
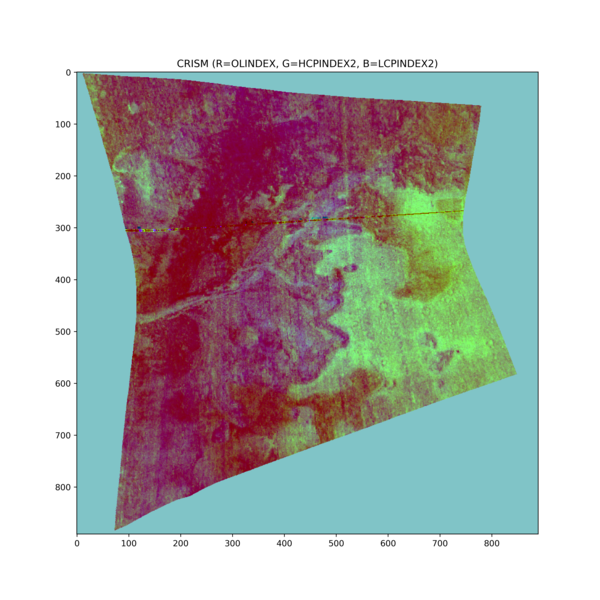
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, Mars
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, MarsThis figure shows an example mineral parameter map image generated using PyHAT. The area in this Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) image is Jezero crater, the landing site of NASA's Mars Perseverance rover.
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, Mars
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, MarsThis figure shows an example mineral parameter map image generated using PyHAT. The area in this Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) image is Jezero crater, the landing site of NASA's Mars Perseverance rover.
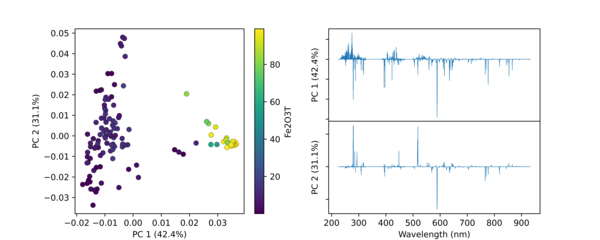
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.
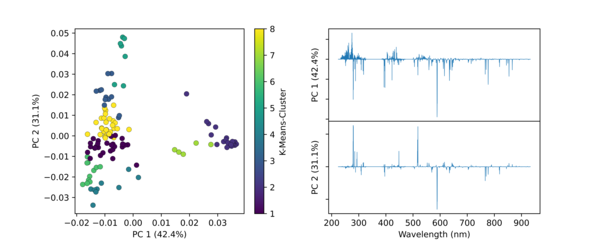
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.
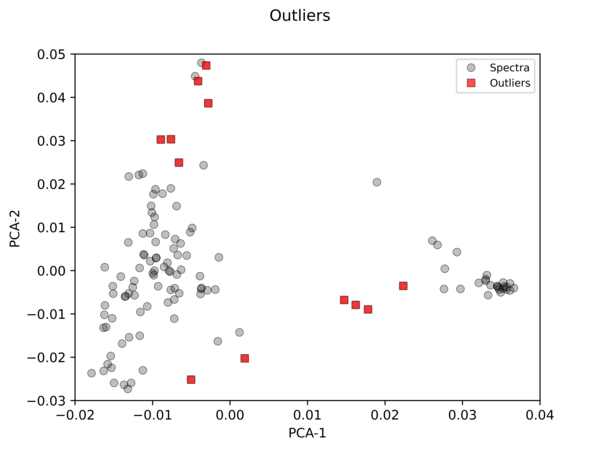
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification Example
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification ExampleThis figure shows an example of outlier identification using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum. Dimensionality was then reduced using principal components analysis (PCA).
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification Example
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification ExampleThis figure shows an example of outlier identification using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum. Dimensionality was then reduced using principal components analysis (PCA).
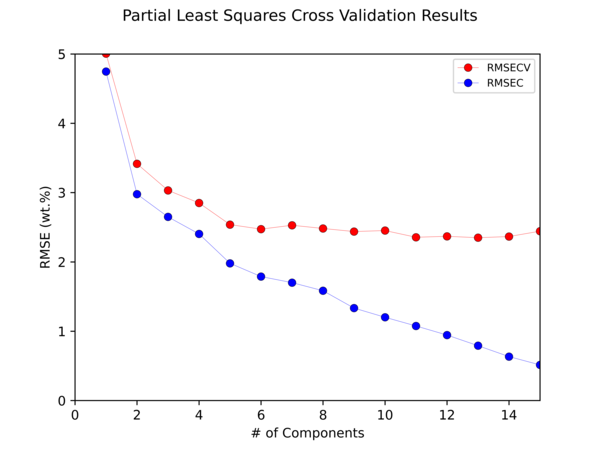
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation Example
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation ExampleThis figure shows the results of cross-validating a Partial Least Squares (PLS) model to predict the abundance of CaO in geologic targets using PyHAT. Cross validation is necessary to optimize the parameters of a regression algorithm to avoid overfitting.
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation Example
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation ExampleThis figure shows the results of cross-validating a Partial Least Squares (PLS) model to predict the abundance of CaO in geologic targets using PyHAT. Cross validation is necessary to optimize the parameters of a regression algorithm to avoid overfitting.
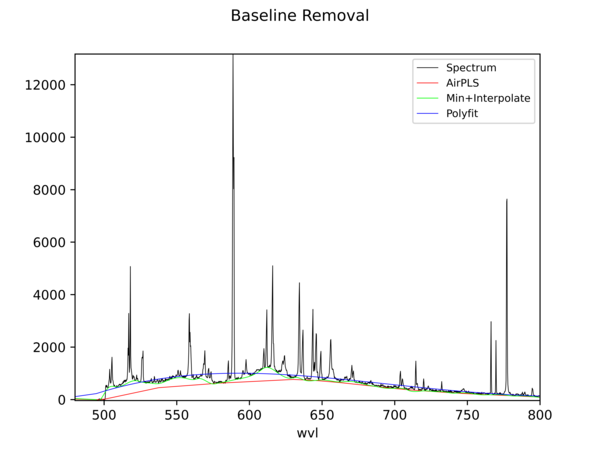
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot ExampleThis figure shows an example spectrum plot generated using PyHAT. The black line is a laser induced breakdown spectroscopy (LIBS) spectrum of a basalt sample. The colored lines show the baseline estimated using several different algorithms.
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot ExampleThis figure shows an example spectrum plot generated using PyHAT. The black line is a laser induced breakdown spectroscopy (LIBS) spectrum of a basalt sample. The colored lines show the baseline estimated using several different algorithms.
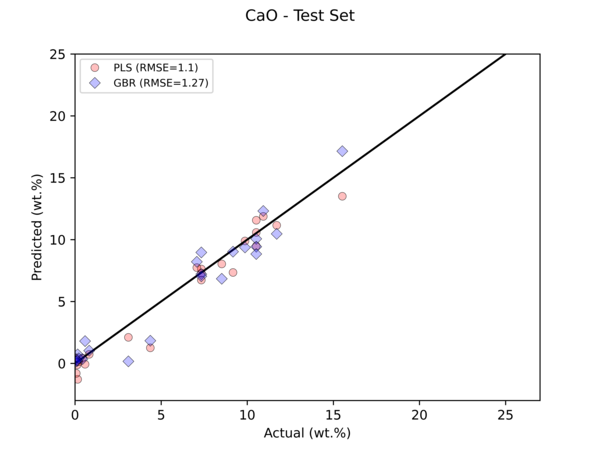
Python Hyperspectral Analysis Tool (PyHAT) Regression Example
Python Hyperspectral Analysis Tool (PyHAT) Regression ExampleThis figure compares the results of two regression models to predict the abundance of CaO in geologic standards based on their laser induced breakdown spectroscopy (LIBS) spectra using PyHAT. The horizontal axis is the independently measured CaO abundance, the vertical axis is the abundance predicted by the models.
Python Hyperspectral Analysis Tool (PyHAT) Regression Example
Python Hyperspectral Analysis Tool (PyHAT) Regression ExampleThis figure compares the results of two regression models to predict the abundance of CaO in geologic standards based on their laser induced breakdown spectroscopy (LIBS) spectra using PyHAT. The horizontal axis is the independently measured CaO abundance, the vertical axis is the abundance predicted by the models.
Python Hyperspectral Analysis Tool (PyHAT) user guide Python Hyperspectral Analysis Tool (PyHAT) user guide
Intense alteration on early Mars revealed by high-aluminum rocks at Jezero Crater Intense alteration on early Mars revealed by high-aluminum rocks at Jezero Crater
Regolith of the crater floor units, Jezero crater, Mars: Textures, composition and implications for provenance Regolith of the crater floor units, Jezero crater, Mars: Textures, composition and implications for provenance
Spatial and temporal distribution of sinuous ridges in southeastern Terra Sabaea and the northern region of Hellas Planitia, Mars Spatial and temporal distribution of sinuous ridges in southeastern Terra Sabaea and the northern region of Hellas Planitia, Mars
Morphology and paleohydrology of intracrater alluvial fans north of Hellas Basin, Mars Morphology and paleohydrology of intracrater alluvial fans north of Hellas Basin, Mars
Overview of the morphology and chemistry of diagenetic features in the clay-rich Glen Torridon Unit of Gale Crater, Mars Overview of the morphology and chemistry of diagenetic features in the clay-rich Glen Torridon Unit of Gale Crater, Mars
In situ recording of Mars soundscape In situ recording of Mars soundscape
Introduction to the Python Hyperspectral Analysis Tool (PyHAT) Introduction to the Python Hyperspectral Analysis Tool (PyHAT)
Post-landing major element quantification using SuperCam laser induced breakdown spectroscopy Post-landing major element quantification using SuperCam laser induced breakdown spectroscopy
Improving ChemCam LIBS long-distance elemental compositions using empirical abundance trends Improving ChemCam LIBS long-distance elemental compositions using empirical abundance trends
Quantification of manganese for ChemCam Mars and laboratory spectra using a multivariate model Quantification of manganese for ChemCam Mars and laboratory spectra using a multivariate model
Alternating wet and dry depositional environments recorded in the stratigraphy of Mt Sharp at Gale Crater, Mars Alternating wet and dry depositional environments recorded in the stratigraphy of Mt Sharp at Gale Crater, Mars
Science and Products
Terrestrial Remote Sensing Data Ingestion with PyHAT (Python Hyperspectral Analysis Tool)
Python Hyperspectral Analysis Tool (PyHAT)
Digital Terrain Models for Mars Sinuous Ridges and Alluvial Fans Projects Digital Terrain Models for Mars Sinuous Ridges and Alluvial Fans Projects
This a version of the logo for the Python Hyperspectral Analysis Tool (PyHAT). It is intended for use in info boxes on the USGS website. The spectrum in the graphic is a laser induced breakdown spectroscopy spectrum, plotted on a logarithmic y axis to emphasize weaker emission peaks.
This a version of the logo for the Python Hyperspectral Analysis Tool (PyHAT). It is intended for use in info boxes on the USGS website. The spectrum in the graphic is a laser induced breakdown spectroscopy spectrum, plotted on a logarithmic y axis to emphasize weaker emission peaks.

Python Hyperspectral Analysis Tool (PyHAT) Data Format Example
Python Hyperspectral Analysis Tool (PyHAT) Data Format ExampleScreenshot showing the simple data format used by the Python Hyperspectral Analysis Tool (PyHAT). Spectra are stored in rows of the table, along with their associated metadata and compositional information.
Python Hyperspectral Analysis Tool (PyHAT) Data Format Example
Python Hyperspectral Analysis Tool (PyHAT) Data Format ExampleScreenshot showing the simple data format used by the Python Hyperspectral Analysis Tool (PyHAT). Spectra are stored in rows of the table, along with their associated metadata and compositional information.

Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, Mars
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, MarsThis figure shows an example mineral parameter map image generated using PyHAT. The area in this Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) image is Jezero crater, the landing site of NASA's Mars Perseverance rover.
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, Mars
Python Hyperspectral Analysis Tool (PyHAT) Mineral Parameter Map Example - Jezero Crater, MarsThis figure shows an example mineral parameter map image generated using PyHAT. The area in this Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) image is Jezero crater, the landing site of NASA's Mars Perseverance rover.

Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis (PCA) Plot ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.

Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering Example
Python Hyperspectral Analysis Tool (PyHAT) Principal Component Analysis K-Means Clustering ExampleThis figure shows an example PCA plot generated using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum.

Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification Example
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification ExampleThis figure shows an example of outlier identification using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum. Dimensionality was then reduced using principal components analysis (PCA).
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification Example
Python Hyperspectral Analysis Tool (PyHAT) Outlier Identification ExampleThis figure shows an example of outlier identification using PyHAT. The input data were laser induced breakdown spectroscopy (LIBS) spectra. PyHAT was used to apply a baseline correction and normalization to the total intensity for each spectrum. Dimensionality was then reduced using principal components analysis (PCA).

Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation Example
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation ExampleThis figure shows the results of cross-validating a Partial Least Squares (PLS) model to predict the abundance of CaO in geologic targets using PyHAT. Cross validation is necessary to optimize the parameters of a regression algorithm to avoid overfitting.
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation Example
Python Hyperspectral Analysis Tool (PyHAT) Partial Least Squares Cross Validation ExampleThis figure shows the results of cross-validating a Partial Least Squares (PLS) model to predict the abundance of CaO in geologic targets using PyHAT. Cross validation is necessary to optimize the parameters of a regression algorithm to avoid overfitting.

Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot ExampleThis figure shows an example spectrum plot generated using PyHAT. The black line is a laser induced breakdown spectroscopy (LIBS) spectrum of a basalt sample. The colored lines show the baseline estimated using several different algorithms.
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot Example
Python Hyperspectral Analysis Tool (PyHAT) Baseline Removal Plot ExampleThis figure shows an example spectrum plot generated using PyHAT. The black line is a laser induced breakdown spectroscopy (LIBS) spectrum of a basalt sample. The colored lines show the baseline estimated using several different algorithms.

Python Hyperspectral Analysis Tool (PyHAT) Regression Example
Python Hyperspectral Analysis Tool (PyHAT) Regression ExampleThis figure compares the results of two regression models to predict the abundance of CaO in geologic standards based on their laser induced breakdown spectroscopy (LIBS) spectra using PyHAT. The horizontal axis is the independently measured CaO abundance, the vertical axis is the abundance predicted by the models.
Python Hyperspectral Analysis Tool (PyHAT) Regression Example
Python Hyperspectral Analysis Tool (PyHAT) Regression ExampleThis figure compares the results of two regression models to predict the abundance of CaO in geologic standards based on their laser induced breakdown spectroscopy (LIBS) spectra using PyHAT. The horizontal axis is the independently measured CaO abundance, the vertical axis is the abundance predicted by the models.



